

Long, intervening noncoding (linc) RNA that can be found in evolutionarily conserved, intergenic regions. Pseudogene that can contain introns since produced by gene duplication.Īnnotated on an artifactual region of the genome assembly. Pseudogene that has mass spec data suggesting that it is also translated.Ī species-specific unprocessed pseudogene without a parent gene, as it has an active orthologue in another species. Pseudogene where protein homology or genomic structure indicates a pseudogene, but the presence of locus-specific Pseudogene owing to a reverse transcribed and re-inserted sequence.
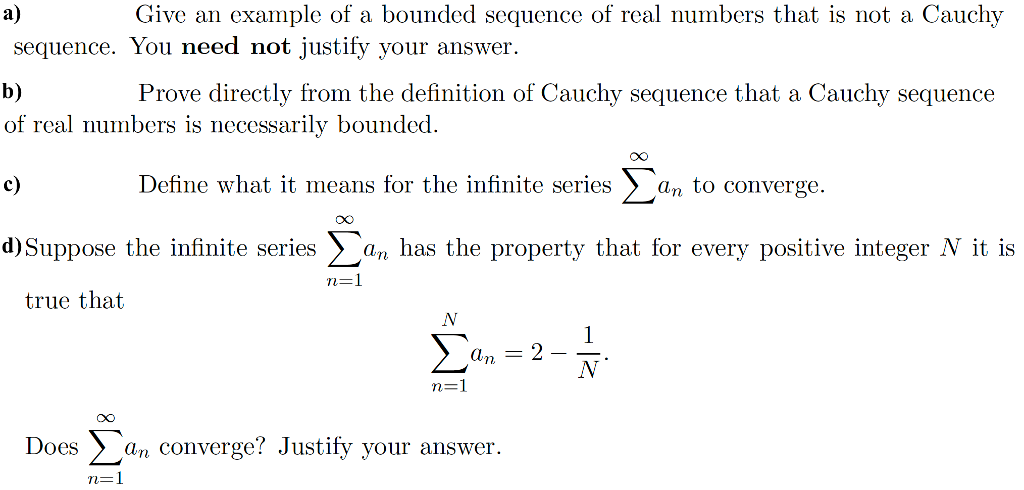
Pseudogene owing to a SNP/DIP but in other individuals/haplotypes/strains the gene is translated. Pseudogene that lack introns and is thought to arise from reverse transcription of mRNA followed by reinsertion of DNA Can be further classified as one of the following. Sometimes these entries have an intact coding sequence or an open but truncated ORF, in whichĬase there is other evidence used (for example genomic polyA stretches at the 3' end) to classify them as a Have homology to proteins but generally suffer from a disrupted coding sequence and an active homologous gene can beįound at another locus. exon or introns) of a protein-coding locus on the opposite strand. Has transcripts that overlap the genomic span (i.e. Long non-coding transcript that contains a coding gene in its intron on the same strand. Long non-coding transcript in introns of a coding gene that does not overlap any exons. Transcript believed to be protein coding, but with more than one possible open reading frame. Transcript which is known from the literature to not be protein coding. Replaces the polymorphic_pseudogene transcript biotype. Not translated in the reference genome owing to a SNP/DIP but in other individuals/haplotypes/strains the transcript is translated. These transcripts are subject to degradation.Īlternatively spliced transcript believed to contain intronic sequence relative to other, coding, variants. Tail attached directly to the CDS without 3' UTR. Transcript that has polyA features (including signal) without a prior stop codon in the CDS, i.e. no matter what the exon structure of the missing portion is the transcript will be
#Sequence definition full
If the variant does not cover the full reference coding sequence then it is annotated as If the coding sequence (following the appropriate reference) of a transcript finishes >50bp from a downstream splice This category hasīeen specifically created for the ENCODE project to highlight regions that could indicate the presence of proteinĬoding genes that require experimental validation, either by 5' RACE or RT-PCR to extend the transcripts, or byĬonfirming expression of the putatively-encoded peptide with specific antibodies. This is used for non-spliced EST clusters that have polyA features. Non-coding RNA predicted to be pseudogene by the Ensembl pipeline Generic long non-coding RNA biotype that replaced the following biotypes: 3prime_overlapping_ncRNA,Īntisense, bidirectional_promoter_lncRNA, lincRNA, macro_lncRNA, non_coding, processed_transcript, Non-coding RNA predicted using sequences from Rfam and miRBase Immunoglobulin (Ig) variable chain and T-cell receptor (TcR) genes imported or annotated according to the IMGT. Please also compare to the VEGAdescriptions.įurther details about the annotation of non-coding RNAs are listed on this Ensembl page. The energy is carried by either radiation or convection, with the latter occurring in regions with steeper temperature gradients, higher opacity or both.Gene/Transcript Biotypes in GENCODE & Ensembl

Energy generated at the core makes its way to the surface and is radiated away at the photosphere. The strong dependence of the rate of energy generation in the core on the temperature and pressure helps to sustain this balance. All main-sequence stars are in hydrostatic equilibrium, where outward thermal pressure from the hot core is balanced by the inward gravitational pressure from the overlying layers.

During this stage of the star's lifetime, it is located along the main sequence at a position determined primarily by its mass, but also based upon its chemical composition and other factors. Stars on this band are known as main-sequence stars or "dwarf" stars.Īfter a star has formed, it creates energy at the hot, dense core region through the nuclear fusion of hydrogen atoms into helium. These color-magnitude plots are known as Hertzsprung–Russell diagrams after their co-developers, Ejnar Hertzsprung and Henry Norris Russell. In astronomy the main sequence is a continuous and distinctive band of stars that appears on plots of stellar color versus brightness. Freebase (0.00 / 0 votes) Rate this definition:


 0 kommentar(er)
0 kommentar(er)
